Output Files
The PATH_TO_OUTPUT_DIR folder contains a logs and work folder for troubleshooting, but for most purposes all the outputs of interest will be in a subfolder called hippunfold with the following structure:
hippunfold/
└── sub-{subject}
├── anat
├── coords
├── qc
├── surf
└── warps
Briefly, anat contains preprocessed volumetric input images and output segmentations in nifti format, surf contains surface data in gifti format, coords contain Laplace fields spanning the hippocampus, warps contains transformations between unfolded and native or ‘unfolded’ space, and qc contains snapshots and useful diagnostic information for quality control.
anat
This folder contains input anatomical images that have been non-uniformity corrected,
motion-corrected, and, where appropriate, averaged and registered.
In this example, a T1w image was used as a standard reference image, but a T2w was also registered and used in tissue segemntation:
sub-001
└── anat
├── sub-001_desc-preproc_T1w.nii.gz
├── sub-001_space-T1w_desc-preproc_T2w.nii.gz
├── sub-001_hemi-R_space-T1w_desc-subfields_atlas-bigbrain_dseg.nii.gz
├── sub-001_hemi-R_space-cropT1w_desc-preproc_T2w.nii.gz
└── sub-001_hemi-R_space-cropT1w_desc-subfields_atlas-bigbrain_dseg.nii.gz
As per BIDS guidelines, desc-preproc refers to preprocessed input images, space-T1w refers to the volume to which the image is registered, hemi refers to the left or right hemisphere (only shown for the right in this example), and dseg (discrete-segmentation) images with desc-subfields contains subfield labels (coded as integers as described in the included volumes.tsv file). The subfield atlas used will also be included, by default as atlas-bigbrain. Note that HippUnfold does most intermediate processing in an unshown (available in the work/ folder) space-corobl which is cropped, upsampled, and rotated. Downsampling to the original T1w space can thus degrade the results and so they are also provided in a higher resolution space-cropT1w space which is ideal for conducting volumetry or morphometry measures with high precision and detail.
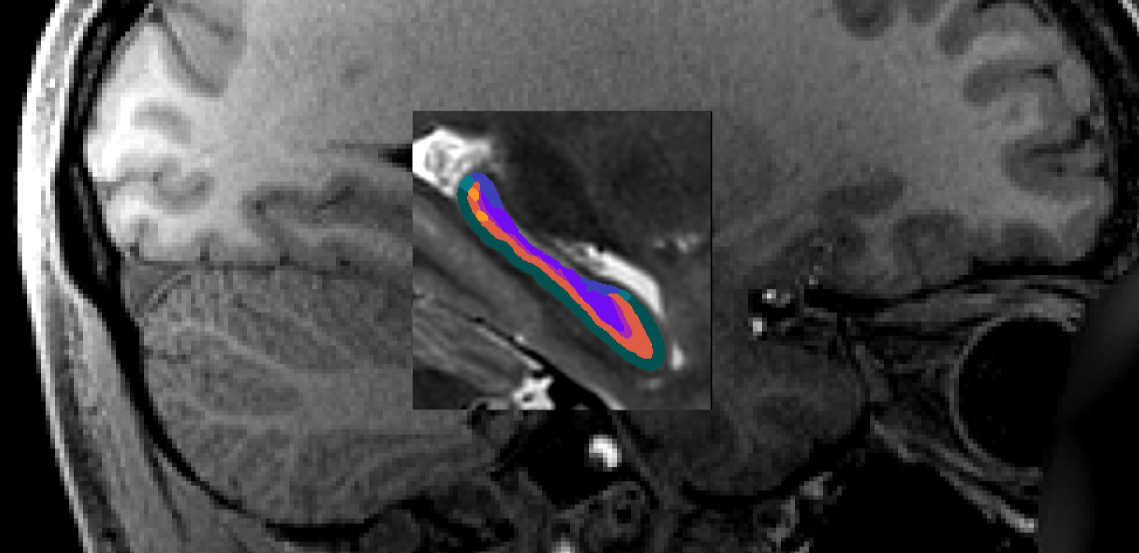
For example, the following image shows a whole-brain T1w image, a
space-cropT1w overlay of the upsampled T2w image (centre square), and a similarly upsampled output
subfield segmentation (colour).
surf
surface meshes
Surface meshes (geometry files) are in .surf.gii format, and are
provided in both the native space (space-T1w) and the unfolded space
(space-unfolded). In each space, there are inner, midthickness,
and outer surfaces, which correspond to white, midthickness, and
pial for cortical surfaces:
sub-{subject}
└── surf
└── sub-001_hemi-R_space-{T1w,unfolded}_den-0p5mm_{inner,midthickness,outer}.surf.gii
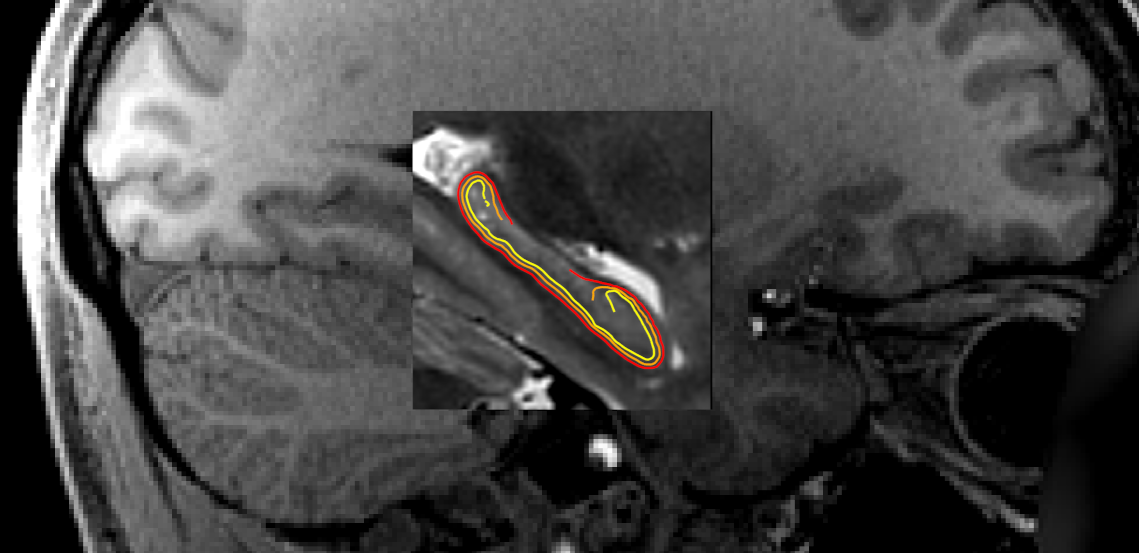
The following shows surfaces inner, midthickness, and outer in
yellow, orange, and red, respectively.
surface densities
Surfaces are provided in different density configurations, and are
labelled based on the approximate vertex spacing in each. The default
density is 0p5mm, which has an approximate vertex spacing of 0.5mm. There are also
1mm and 2mm surfaces which have 1mm or 2mm spacing, respectively (suitable for
lower-resolution BOLD data). Previous versions of hippunfold exclusively
used a unfoldiso template surface, formed by a 254x126 grid in
the unfolded space, however a downside of this template is that it
results in very non-uniform vertex spacing when transformed to the
native space. The newer 0p5mm,
1mm and 2mm surfaces are designed to have
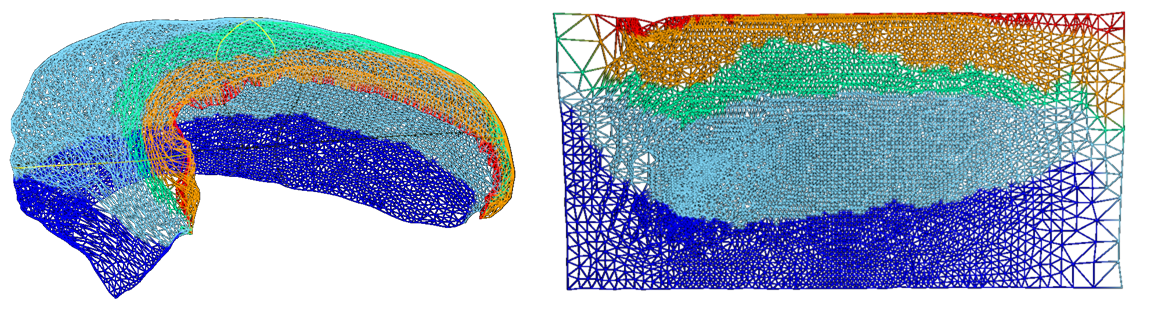
closer to uniform vertex spacing in native space, though vertex spacing will not remain uniform when unfolded.
This is illustrated in the the following den-1mm mesh in folded and
unfolded space.
All surfaces of the same density (e.g. 1mm), in both
space-T1w and space-unfolded, share the same mesh topology and have
corresponding vertices with each other. The vertex locations for
unfolded surfaces are identical for all subjects as well (note that this
of course is not the case for the space-T1w surfaces).
surface metrics
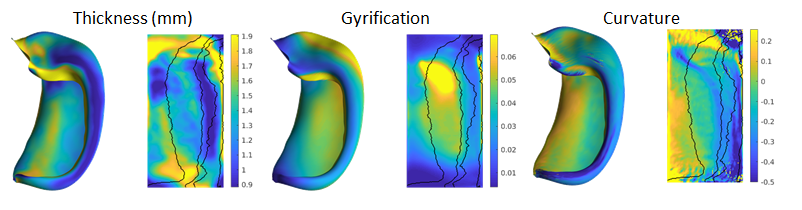
In addition to the geometry files, surface-based shape metrics are
provided in .shape.gii format. The thickness, curvature and surface
area are computed using the same methods as cortical surfaces, based on
the surface geometry files, and are provided in the T1w space. The
gyrification metric is the ratio of native to unfolded surface area, or
equivalently, the scaling or distortion factor when unfolding:
sub-{subject}
└── surf
└── sub-001_hemi-{L,R}_space-T1w_den-0p5mm_label-hipp_{thickness,curvature,gyrification}.shape.gii
└── sub-001_hemi-{L,R}_space-T1w_den-0p5mm_label-dentate_{curvature,gyrification}.shape.gii
These metrics are shown in both folded and unfolded space in the images below. Note that these results are from group-averaged data and so individual subject maps may show considerably more variability.
surface labels
The subfield labels from unfolded atlases are also provided for each
subject, in .label.gii format. Analogous to the volume-based labels,
the name of the atlas (default: bigbrain) is in the file name.
sub-{subject}
└── surf
└── sub-001_hemi-{L,R}_space-T1w_den-0p5mm_label-hipp_atlas-bigbrain_subfields.label.gii
cifti files
In addition to lateralized .shape.gii and .label.gii metrics and labels,
we also provide data mapped to hippocampi from hemispheres in a single
file using the corresponding CIFTI formats, .dscalar.nii and .dlabel.nii.
Note: since CIFTI does not support hippocampus
surfaces (yet), we make use of the CORTEX_LEFT and CORTEX_RIGHT labels for
the hippocampal surfaces.
sub-{subject}
└── surf
├── sub-001_space-T1w_den-0p5mm_label-{hipp,dentate}_{thickness,curvature,gyrification}.dscalar.nii
└── sub-001_space-T1w_den-0p5mm_label-hipp_atlas-bigbrain_subfields.dlabel.nii
spec files
Finally, these files are packaged together for easy viewing in
Connectome Workbench, wb_view, in the following .spec files, for
each hemisphere and structure separately, and combined:
sub-{subject}
└── surf
├── sub-001_hemi-{L,R}_space-T1w_den-0p5mm_label-{hipp,dentate}_surfaces.spec
└── sub-001_space-T1w_den-0p5mm_label-{hipp,dentate}_surfaces.spec
New: label-dentate
HippUnfold v1.0.0 introduces label-dentate files which represent a distinct surface making up the dentate gyrus (reflecting its distinct topology from the rest of the cortex). The rest of the surfaces are given the name label-hipp to differentiate them from these new files.
These are illustrated in the following image (orange represents the usual hippocampal midthickness surface, while violet shows the new dentate surface):
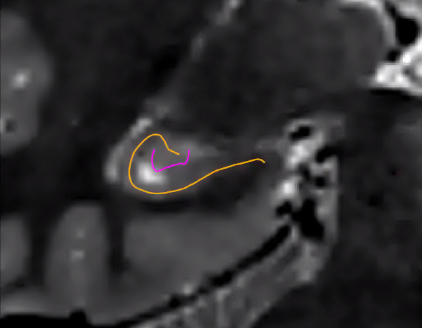
Note that the dentate uses the same unfolding methods as the rest of the hippocampus, but with several caveats. Given its small size, its boundaries are not easily deliminated and so inner, outer, and thickness gifti surfaces are omitted. Furthermore, Laplace coordinates and therefore vertex spacing are not guaranteed to be topologically equivalent as they are obtained through volumetric registration with the template shape injection step of this workflow.
Corresponding coords and warp files are also generated.
New: myelin maps
If your dataset has T1w and T2w images (and you are using --modality=T1w or --modality=T2w), then you can enable the generation of myelin maps as the ratio of T1w over T2w images. This division is done in the corobl space, and provides myelin.shape.gii surface metrics, and also includes these in the CIFTI and spec files.
This option is enabled with the --generate-myelin-maps command-line option.
coords
Hippunfold also provides images that represent anatomical gradients
along the 3 principal axes of the hippocampus, longitudinal from
anterior to posterior, lamellar from proximal (dentate gyrus) to distal
(subiculum), and laminar from inner (SRLM) to outer. These are provided
in the images suffixed with coords.nii.gz with the direction indicated
by dir-{direction} as AP, PD or IO, and intensities from 0 to 1,
e.g. 0 representing the Anterior end and 1 the Posterior end.
Here is an example showing coronal slices of the hippocampus with the PD, IO, and AP (sagittal slice) overlaid.
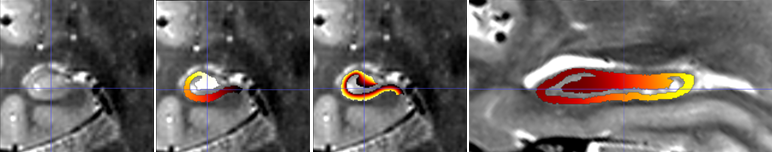
Note that these images have been resampled to space-corobl which is
the space in which most processing is done internally. These can be seen
in the work/ output directory or specified as a possible output space.
warps
ITK transforms to warp images between the T1w space to the unfolded
space, are provided for each hippocampus:
sub-{subject}
└── seg
└── sub-001_hemi-R_from-T1w_to-unfold_mode-image_xfm.nii.gz
└── sub-001_hemi-R_from-unfold_to-T1w_mode-image_xfm.nii.gz
These are ITK transforms that can transform any image that is in T1w
space (can be any resolution and FOV, as long as aligned to T1w), to
the unfolded hippocampal volume space, and vice-versa. You can use the
warp itself as a reference image, e.g.:
antsApplyTransforms -d 3 \
-i sub-001_space-T1w_FA.nii.gz \
-o sub-001_hemi-L_space-unfolded_FA.nii.gz \
-t sub-001_hemi-L_from-T1w_to-unfold_mode-image_xfm.nii.gz \
-r sub-001_space-unfolded_refvol.nii.gz
Additional Files
The top-level PATH_TO_OUTPUT_DIR contains additional folders:
├── hippunfold
├── logs
├── work
└── .snakemake
The hidden .snakemake folder contains a record of the code and parameters used,
and paths to the inputs.
Workflow steps that write logs to file are stored in the logs
subfolder, with file names based on the rule wildcards (e.g. subject,
hemi, etc..).
Intermediate files are stored in the work folder. These files and
folders, similar to results, are generally named according to BIDS. This
folder will have tar.gz files for each subject, unless the
--keep_work option is used.
If the app is run in workflow mode
(--workflow-mode/-W) which enables direct
use of the snakemake CLI to run hippunfold, the
hippunfold and work folders will be placed
in a results folder.