Full Documentation: here
Hippunfold
This tool aims to automatically model the topological folding structure
of the human hippocampus, and computationally unfold it.
This is especially useful for:
Visualization
Topologically-constrained intersubject registration
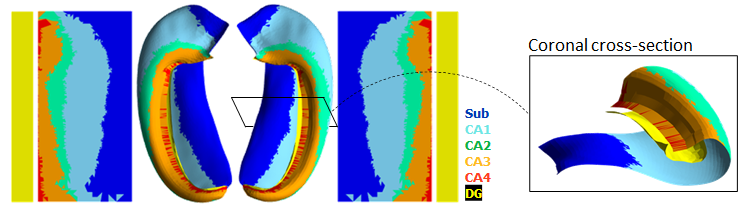
Parcellation (ie. registration to an unfolded atlas)
Morphometry (eg. thickness, surface area, curvature, and gyrification measures)
Quantitative mapping (eg. map your qT1 MRI data to a midthickness surface; extract laminar profiles perpendicular to this surface)
NEW: Version 1.3.x release
Major changes include the addition of unfolded space registration to a reference atlas harmonized across seven ground-truth histology samples. This method allows shifting in unfolded space, providing even better intersubject alignment.
Note: this replaces the default workflow, however you can revert to the legacy workflow, disabling unfolded space registration, by setting --atlas bigbrain or --no-unfolded-reg
Read more in our manuscript
Also the ability to specify a new experimental UNet model that is contrast-agnostic using synthseg and trained using more detailed segmentations. This generally produces more detailed results but has not been extensively tested yet.
Note: Docker containers for version 1.3.x and above do not come pre-shipped with nnU-net models (and are accordingly more lightweight!) - models are downloaded automatically when running, but please see the FAQ for more information!
Workflow
The overall workflow can be summarized in the following steps:
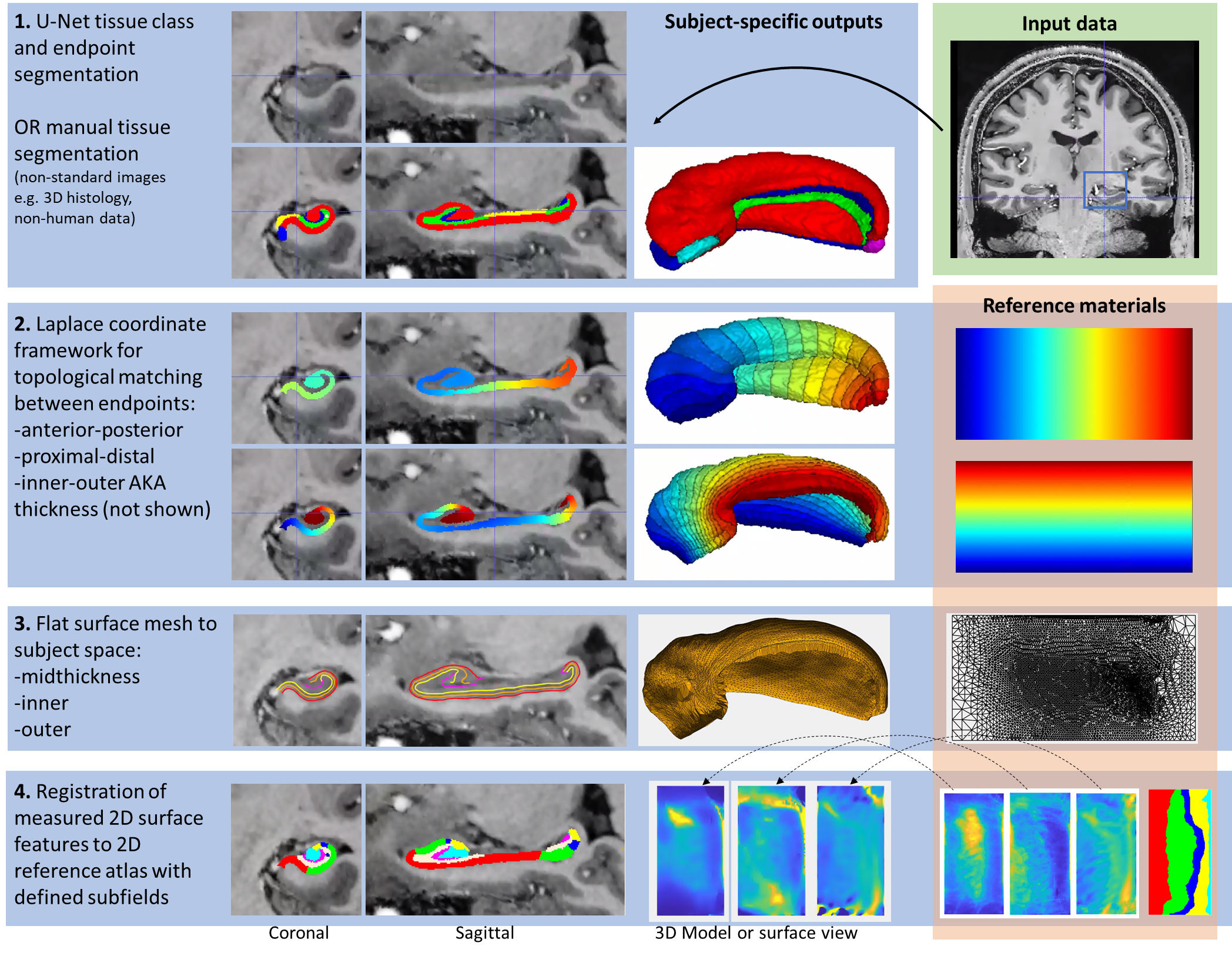
For more information, see Full Documentation: here
Additional tools
For plotting, mapping fMRI, DWI or other data, and manipulating surfaces, see here
For statistical testing (spin tests) in unfolded space, see here
Publications
HippUnfold methods paper
DeKraker, J., Haast, R. A., Yousif, M. D., Karat, B., Lau, J. C., Köhler, S., & Khan, A. R. (2022). Automated hippocampal unfolding for morphometry and subfield segmentation with HippUnfold. Elife, 11, e77945. link
Please cite this if you use any version of HippUnfold)
Unfolded space registration and multihist7 atlas
DeKraker Jordan, Palomero-Gallagher Nicola, Kedo Olga, Ladbon-Bernasconi Neda, Muenzing Sascha E.A., Axer Markus, Amunts Katrin, Khan Ali R., Bernhardt Boris, Evans Alan C. (2023) Evaluation of surface-based hippocampal registration using ground-truth subfield definitions eLife 12:RP88404 link
Please cite this if you use HippUnfold version >= 1.3.0)
Commentary on surface-based hippocampal segmentation
DeKraker J, Köhler S, Khan AR. Surface-based hippocampal subfield segmentation. Trends Neurosci. 2021 Nov;44(11):856-863. doi: 10.1016/j.tins.2021.06.005. Epub 2021 Jul 22. PMID: 34304910. link

